Data preparation
The “Rules of Thumb” for Data Preparation
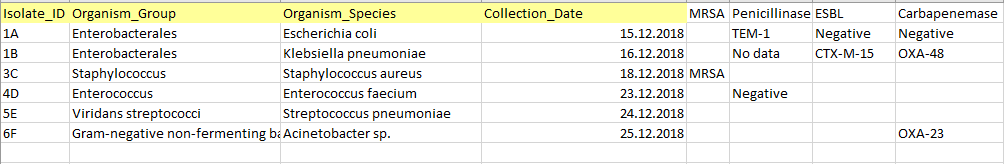
Accurate, well-formated and structured source data is essential for successful analysis using the AMRcloud platform.
- The source data table should include individual isolate records and NOT aggregated statistics. Each isolate record should be represented by a separate row in the table.
- All AST Data and Metadata should be organized in columns with each column representing only a single type of data (number, date, character string).
- The Metadata must include four mandatory columns: Isolate Identifier, Organism/Species Name, Organism Group, and Date. All other metadata (e.g. geographical data, resistance markers, etc.) is optional.
- The AST Data may be of three different types: minimum inhibitory concentrations (MICs), inhibition zone diameters, and susceptibility categories (S/I/R). Headers of the columns representing AST Data should contain a generic name of antimicrobial agent and one of the three suffixes ("_mic”, “_dd” or “_sir”) denoting each type of data.
Example Data
The basic requirements for the source data table are listed in the “Read Me” tab of the data import wizard. An example data file for use as a reference for data formatting can be downloaded from a link at the bottom of the “Read Me” tab.
Basic Requirements
The data should be in the required format to be handled by AMRcloud.
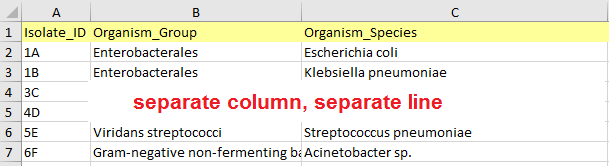
- The data table should be flat (i.e. it should contain columns and rows with no additional separators, no hierarchical structures).
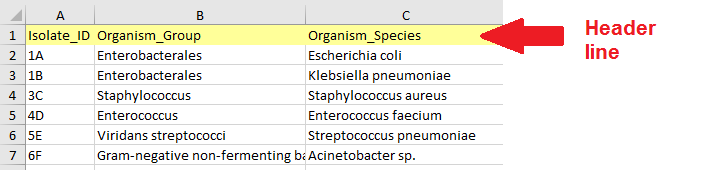
Data table structure - The first row of the table must be a header row with column names.
Header row - The table should contain four mandatory metadata columns (although the column names may be different):
- Isolate Identifier (ID)
- Organism/Species Name
- Organism Group
- Date
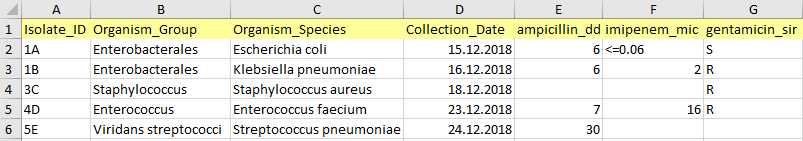
Isolate ID
Isolate ID can be any combination of alphanumeric characters, except special symbols (?,!,%$#><), and can be non-unique. Records (rows) with empty ID fields are not imported.
Organism/Species Name
Only the Latin binomial names should be used for microbial species (see the official nomenclature https://lpsn.dsmz.de). Abbreviations in the species names are not allowed. Proper genus and species names are automatically recognized to determine susceptibility categories to antimicrobial agents using specific breakpoints.
Right:
Staphylococcus aureus, Streptococcus pneumoniae
Wrong:
S. aureus, S. pneumoniae
Organism Group
Any taxonomic or non-taxonomic, full or abbreviated names can be used to define Organism Groups, however, the names should be consistent throughout the column. Grouping of organisms (species) is used for analyzing combined statistics (e.g. resistance prevalence) of several species.
Right:
Enterobacterales, Staphylococci, GN anaerobes, Enterobacterales, Staphylococci, GN anaerobes (all names are standardized)
Wrong:
Enterobact, Staph, Entbact, Staphylococci, GN anaerobes, Gram-neg anaerobes (names are not standardized)
Date
The import wizard can automatically detect and interpret various Date formats, however, the Date format must be consistent throughout the column. Records without a Date are not imported.
Right:
12.08.2018, 20.05.2017, 16.03.2014 (same format DD.MM.YYYY)
Wrong:
13.08.2018, 20/05/19, 03/16/2014 (inconsistent format DD.MM.YYYY, DD/MM/YYYY, MM/DD/YYYY)
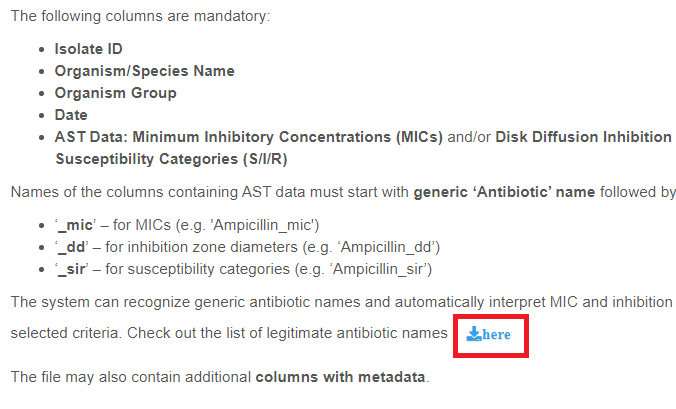
AST Data
The following types of AST Data can be handled by AMRcloud:
- Minimum Inhibitory Concentrations (MICs)
- Disk Diffusion Inhibition Zone Diameters
- Susceptibility Categories (S/I/R)
You can provide an unlimited number of columns with AST Data for various antibiotics and can include data of different types for the same antibiotic in different columns.
Please strictly follow the rules below for naming the columns and choosing the format of AST Data entries.
The header of the columns containing AST Data must start with full generic name of antibiotic in English followed by the underscore symbol and must end with one of the three suffixes denoting the type of data:
| Minimum Inhibitory Concentrations | _mic |
| Disk Diffusion Inhibition Zone Diameters | _dd |
| Susceptibility Categories | _sir |
Example:
tobramycin_sir, tetracycline_sir, amoxicillin-clavulanic acid_dd, vancomycin_mic
The list of legitimate antibiotic names can be downloaded from the link at the “Read Me” tab of the data import wizard. The quantitative AST Data (MIC and inhibition zone values) for the known antibiotics are automatically interpreted into susceptibility categories using selected criteria (breakpoints).
Other (arbitrary) names of antimicrobial agents can be used in the column headers, however, for such agents, quantitative AST Data are not translated into susceptibility categories unless you define custom criteria (breacpoints) as explained in Step 4. If you want to use alternative breakpoints for certain antimicrobial agents, you can modify their names as shown in the example below:
florfenicol_veterinary_mic, marbofloxacin_veterinary_dd
It is allowed to indicate the load of the disk between the name of the antibiotic and the suffix _dd:
amoxicillin-clavulanic acid_2-1_dd, moxifloxacin_5_dd
The requirements for the format of AST Data entries are as follows:
-
Minimum Inhibitory Concentration Values
- MIC values are measured in mg/L.
- Values must be numeric.
- Non-numeric values containing expressions “<=”, “>” or “>=” are automatically converted into numeric.
- Symbols “<=” and “>=” preceding the numbers are removed.
- Numbers preceded by “>” symbol are multiplied by two.
-
Disk Diffusion Inhibition Zone Diameter Values
- Inhibition Zone Diameter values are measured in mm.
- Values must be integers greater than or equal to six.
-
Susceptibility Category Values
- The only allowed values are “S”, “I”, and “R”.
- Values such as “S/I” or “I/R” are not allowed.
Additional Metadata
In addition to Mandatory Columns and AST Data, your table may include other optional columns with information about sources of isolates (including geospatial data), patient clinical and demographic data, specific characteristics of isolates (including phenotypic and genotypic resistance markers), etc. These columns will be used as parameters for data selection, filtration, and categorization.
Spatial Information Column(s)
Spatial Information includes geographic objects (geolocations) and, optionally, their coordinates (latitude and longitude). You may provide various types of objects as geolocations: countries, regions, states, natural areas, cities, smaller locality types or even detailed addresses (e.g. hospital buildings). At Step Three of the data import wizard, an automatic geocoding is performed to assign geographic coordinates to the known geolocations. You can also do it manually if automatic geocoding did not work.
If your data table already contains the two columns with latitude and longitude coordinates, you can check the box and then select these columns at Step One of the data import wizard.
Text (String) Metadata Columns
You can import a maximum of twelve parameters (columns) containing text metadata (including the column with geographic names) to a single Data Set.
Numeric Metadata Column
You can import one parameter (column) containing continuous numeric variable such as “age”, “weight”, etc. Please note that if you use a numeric parameter, all cells in the column must be filled in with the number, records (rows) with empty numeric fields are not imported.
Resistance Markers Columns
Resistance Markers may represent any isolate characteristics (phenotypes, complex genotypes, genes or mutations) that are relevant to antibiotic resistance. Markers can be assigned to groups with each Group of Markers set in a separate column. For example, the column with the header “ESBL” may contain the entries like: “CTX-M-15”, “TEM-3”, “SHV-2”, “SHV-2+CTX-M-15”, “ESBL-negative”, etc. You can provide an unlimited number of columns with Resistance Markers. To ensure consistency, the data on Resistance Markers can be presented as follows:
| Results of testing for specific markers | Entries |
|---|---|
| Positive results | "OXA-48", "KPC", "CTX-M-15", "MRSA", "VRE" etc. |
| Negative result(s) | "Negative" |
| Not tested | "No data" |
| Not applicable for a particular organism/species | empty cell |