Link CAESAR and AMRcloud
This tutorial shows you how to:
- prepare and upload CAESAR data table to the AMRcloud online platform.
- convert the data from the AMRcloud online platfrom to CAESAR data-table format.
From CAESAR to AMRcloud
Isolates data in CAESAR are stored in a format with one antibiotic per row.
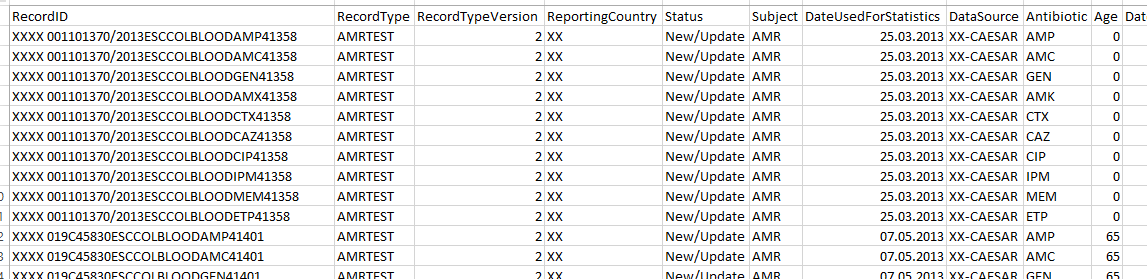
But AMRcloud table has a format with a single row for each isolate and separate columns for each antibiotic.
To convert the CAESAR file to AMRcloud format use the following link https://public.amrcloud.net/caesar.
Uploading a File
Press the Choose file button and select the CAESAR file.
The upload will start automatically. When the upload is complete, the converter checks the file content and previews it on screen.
If the upload is successful, you will see the following:
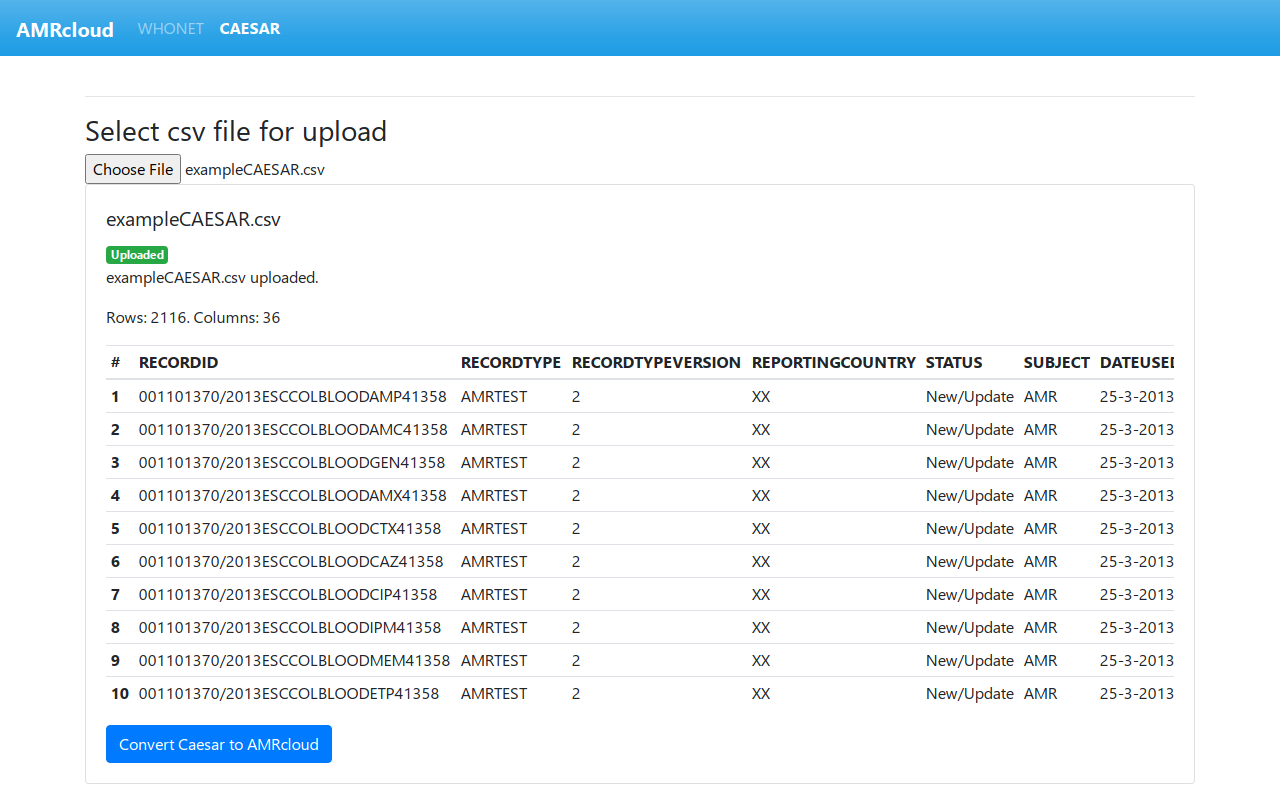
If you see a preview table and the Convert Caesar to AMRcloud button, the file was uploaded correctly.
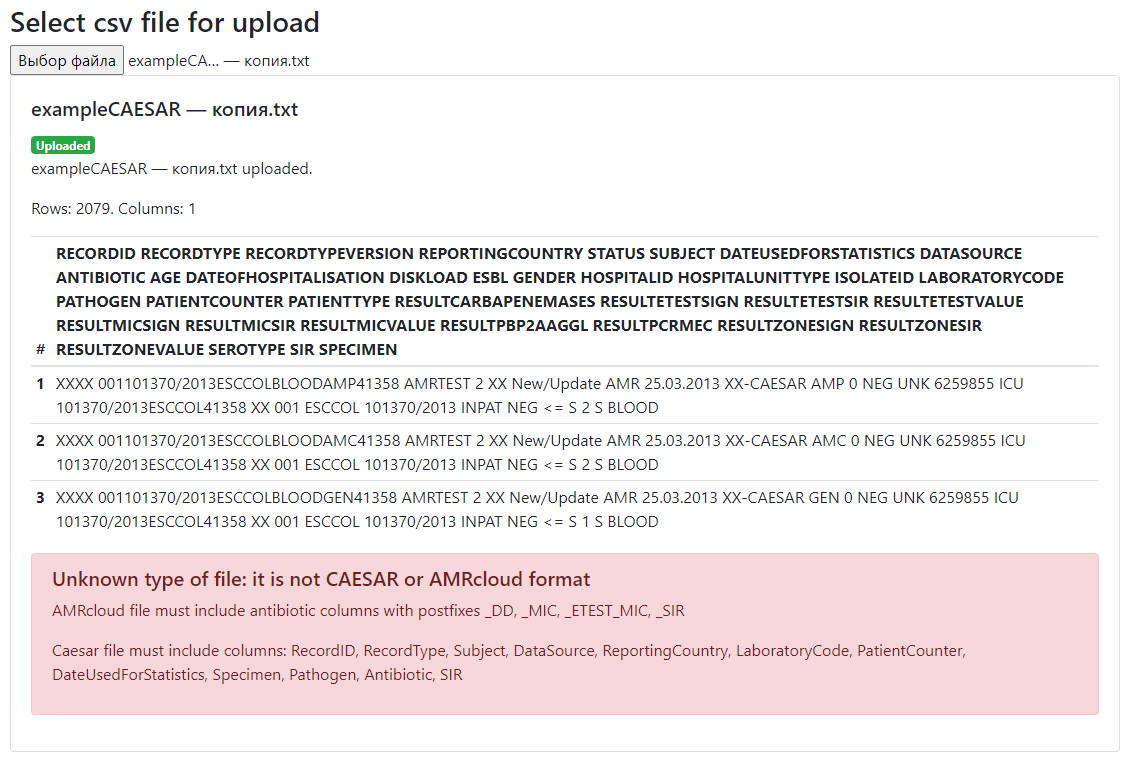
If you see a list of lines (not a table) and an error message, further file processing is not possible. It means that file does not match the specified format. Try to save your gile into CSV format, semicolon delimited, UTF-8 encoding. Another reason for the error may be the absence of required columns in the CAESAR format file: if at least one column is missing, the file is considered invalid.
Converting a File
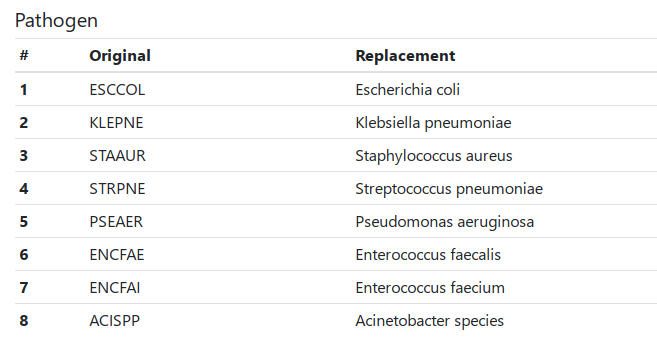
During a conversion process, the table is being spreaded and the CAESAR abbreviated codes for antibiotics (column Antibiotic) and microorganisms (column Pathogen) are being replaced by the corresponding full names and ‘suffix codes’ that can be read by the AMRcloud.
Replacements are preformed according to the CAESAR study protocol. Columns except Antibiotic and Pathogen remain unchanged.
Press the Convert Caesar to AMRcloud button. Once the conversion is complete, a report is displayed.
On the top of the report, there are warnings from the conversion process that provide important information you should pay attention to.
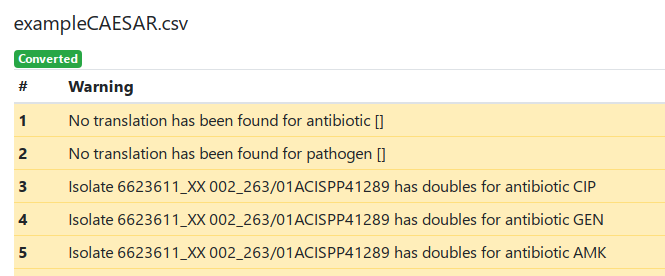
There are three types of warnings:
- No translation has been found for antibiotic []. This message is displayed when a antibitoic CAESAR code is not found in the translation table. In such case, the original name is preserved. If necessary, change the name(s) manually as required after finishing the conversion. In the example shown below, no translation has been found for the empty antibiotic code due to empty lines in the end of file.
- No translation has been found for pathogen []. This message is displayed when a pathogen CAESAR code is not found in the translation table. In such case, the original name is preserved. If necessary, change the name(s) manually as required after finishing the conversion. In the example shown below, no translation has been found for the empty pathogen code due to empty lines in the end of file.
- Isolate has doubles for antibiotic. This message is displayed when one isolate has more than one lines with the same antibiotic name. For the unique isolate ID is used a concatenation of fields HospitalID_LaboratoryCode_IsolateID_PatientID.
A list of translated codes appears below the warnings. Inspect the translations and, if necessary, correct them manually.

Downloading a File
To download the converted file, press the ‘Download’ button. The file should be ready to upload to the AMRcloud without additional modifications, except when you see critical warnings described above.
The import of data files using the AMRcloud Import Wizard is described in the tutorial.
From AMRcloud to CAESAR
Login into your AMRcloud account:
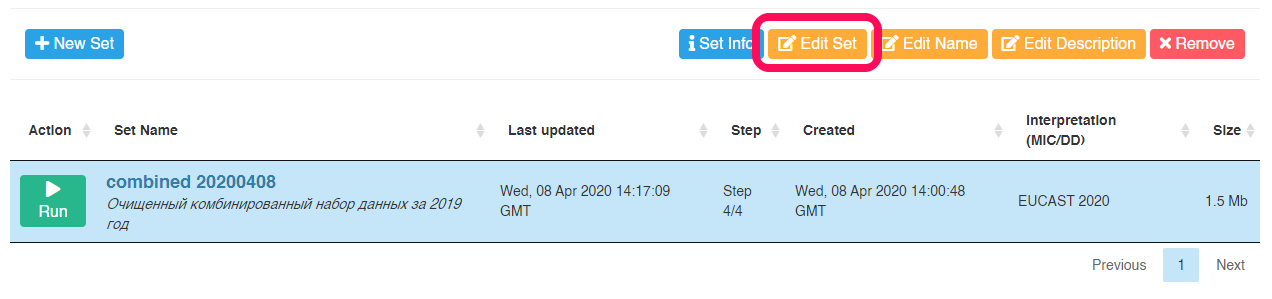
- Choose dataset for export and press the Edit set button;
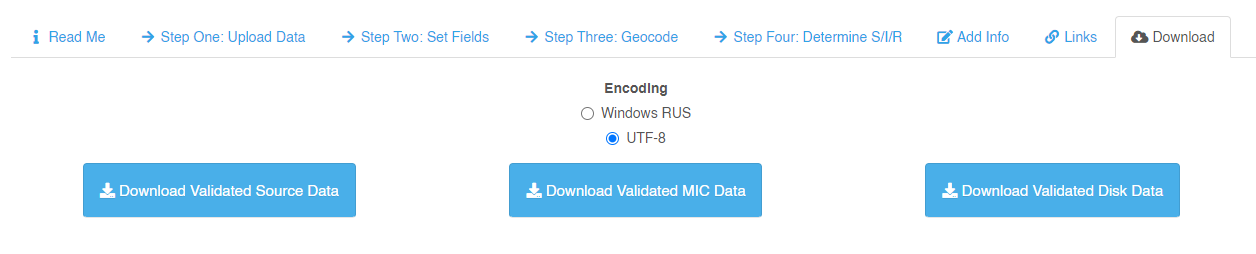
- Go to the Download tab, check the UTF-8 encoding and press the button.
Isolates data in CAESAR are stored in a format with one antibiotic per row, so we need to “collapse” our AMRcloud table.
To convert the AMRcloud file to CAESAR format use the following link https://public.amrcloud.net/caesar.
Uploading a File
Press the Choose file button and select the AMRcloud file.

The upload will start automatically. When the upload is complete, the converter checks the file content and previews it on screen.
If the upload is successful, you will see the following:
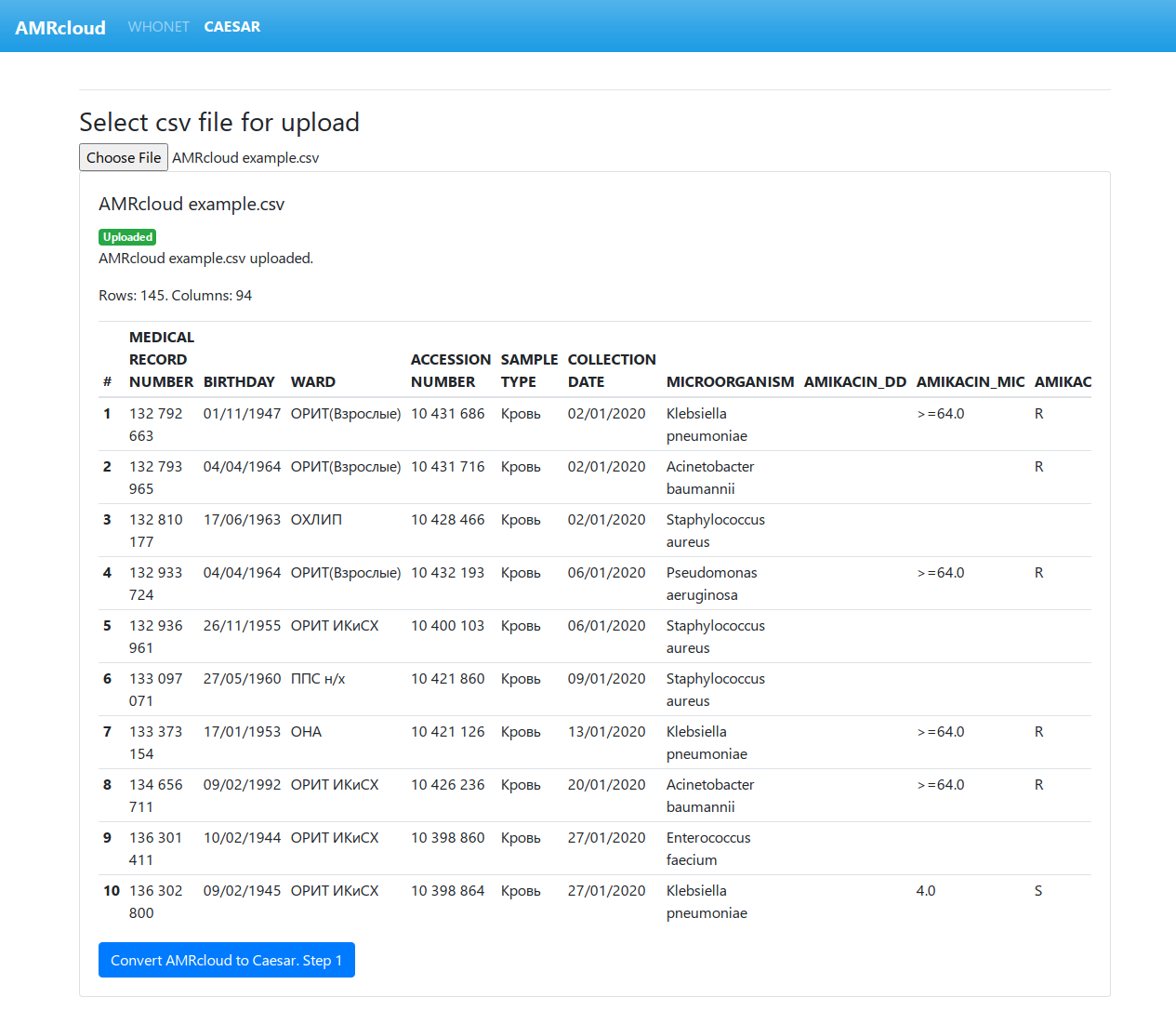
If you see a preview table and the Convert AMRcloud to Caesar. Step 1 button, the file was uploaded correctly.
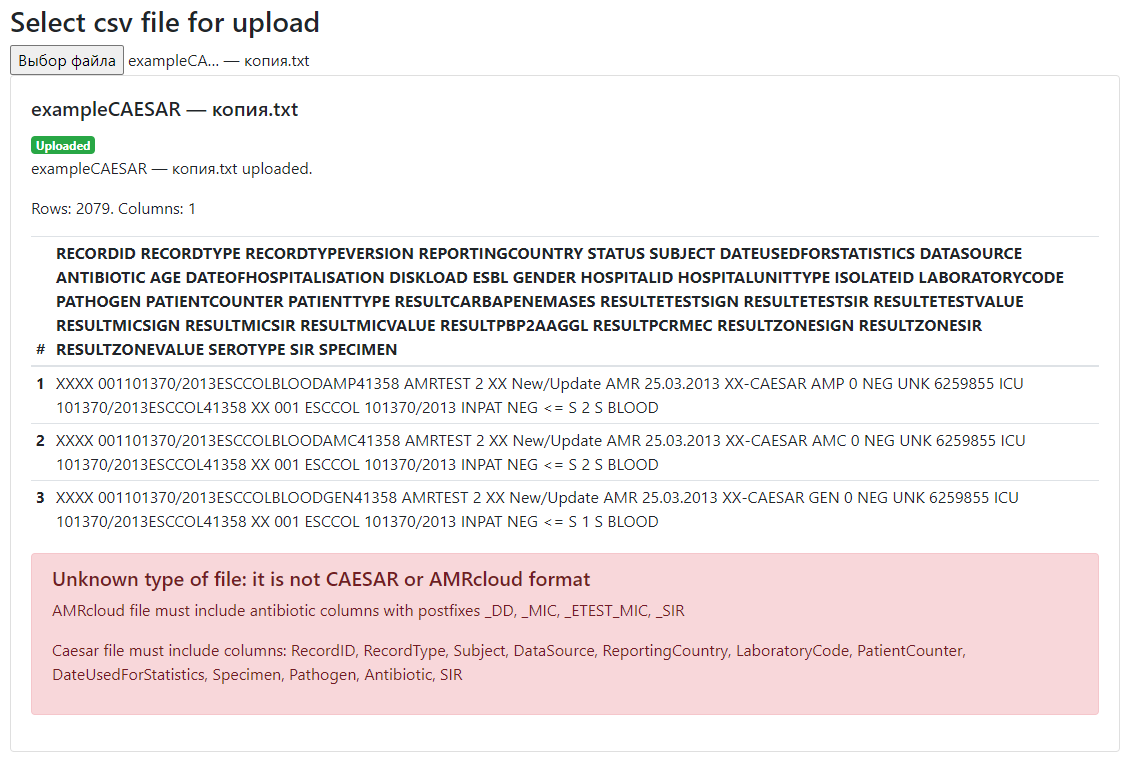
If you see a list of lines (not a table) and an error message, further file processing is not possible. It means that file does not match the specified format. Try to save your gile into CSV format, semicolon delimited, UTF-8 encoding. Another reason for the error may be the absence of required columns in the AMRcloud format file: antibiotics columns with suffixes _dd, _mic, _etest_mic, _sir. The file is considered invalid if there are no antibiotic columns.
Converting a File. Step 1
Before the convertion press the Convert AMRcloud to Caesar. Step 1 button.
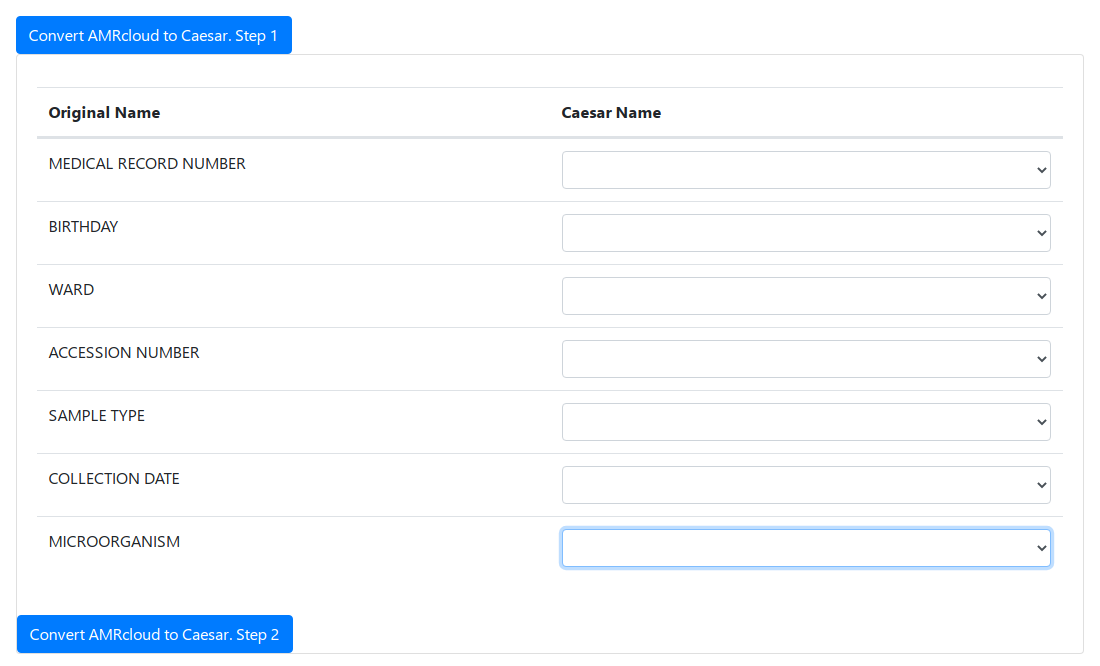
On the first step the fields of the original AMRcloud file have to be matched with the default fields of the CAESAR file. Full description of CAESAR file columns you can find in the CAESAR study protocol.
To match the MICROORGANISM column is required. The rest of the fields are matched at our opinion. If you do not want to match the field, you can leave the drop-down list empty.
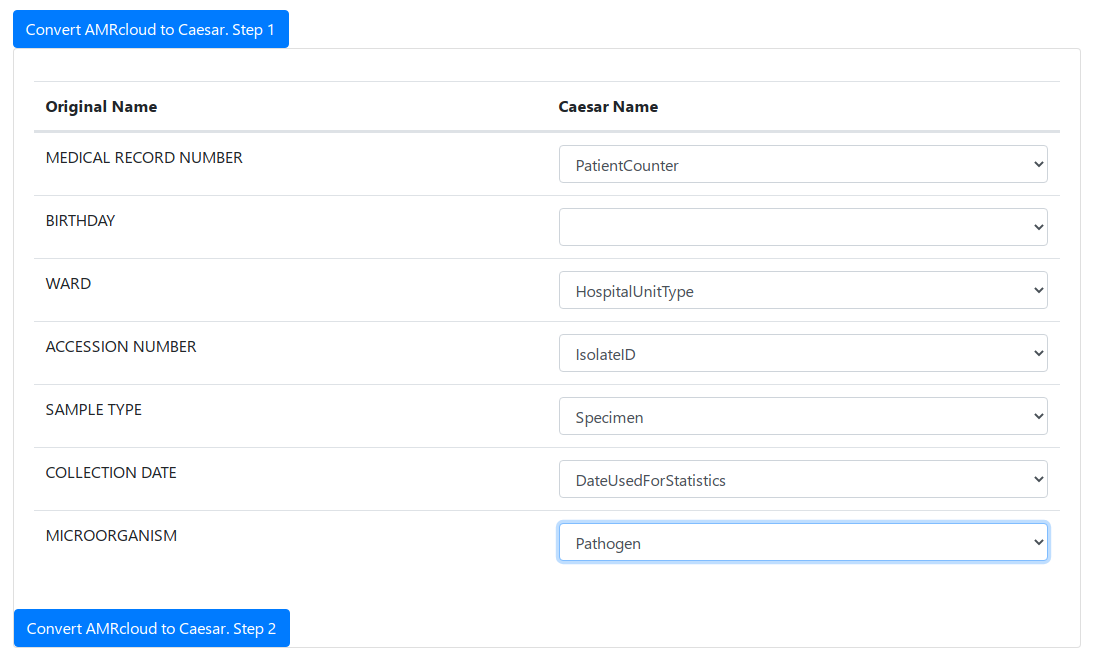
After the fields are matched, you can proceed to step 2.
Converting a File. Step 2
Press the Convert Caesar to AMRcloud button. Once the conversion is complete, a report is displayed.
On the top of the report, there are warnings from the conversion process that provide important information you should pay attention to.
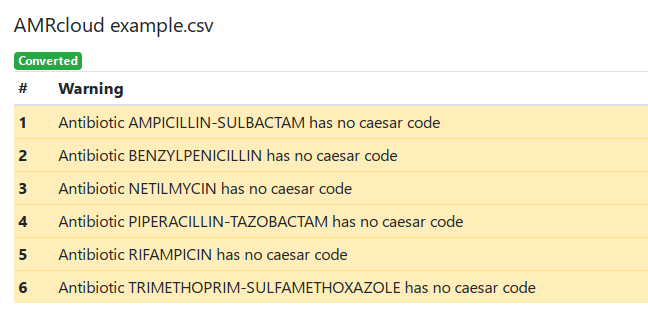
There are three types of warnings:
- No Antibiotic code found for []. This message is displayed when a antibitoic code is not found in the translation table. In such case, the original name is preserved. If necessary, change the name(s) manually as required after finishing the conversion.
- No Specimen code found for []. This message is displayed when a specimen code is not found in the translation table. In such case, the original name is preserved. If necessary, change the name(s) manually as required after finishing the conversion.
- No Pathogen code found for []. This message is displayed when a pathogen code is not found in the translation table. In such case, the original name is preserved. If necessary, change the name(s) manually as required after finishing the conversion.
A list of translated codes appears below the warnings. Inspect the translations and, if necessary, correct them manually.
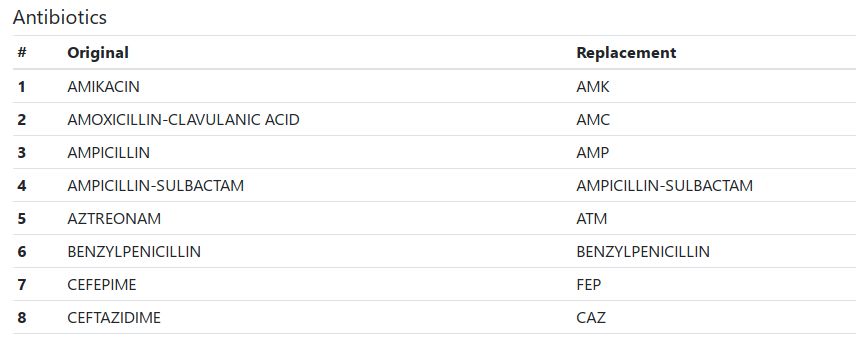

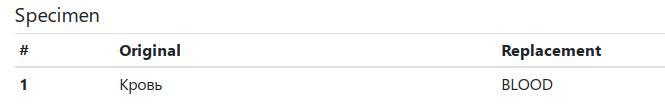
Downloading a File
To download the converted file, press the Download button.