Link Vitek 2 and AMRcloud
This tutorial shows you how to export and convert the data from Vitek 2 system for uploading to the AMRcloud online platform.
The basic steps of uploading data to the AMRcloud online platform are described in the AMRcloud Data Import Tutorial
VITEK 2 export
You can export only isolates that have moved to an inactive area.
The time for isolates to move to the inactive area is set in the general settings. The default is 14 days.
Incomplete isolates (requiring verification or microorganism/patient information) do not move to the inactive area. They must be switched to the status Completed.
In the main window of the program, you should click on the Tools button and select Export inactive isolates.

In the new window, set the date range, check the MIC, Phenotype, Detailed information about the panel. Select other options as needed. Click on the Export button (icon with disk and arrow in the upper right corner).
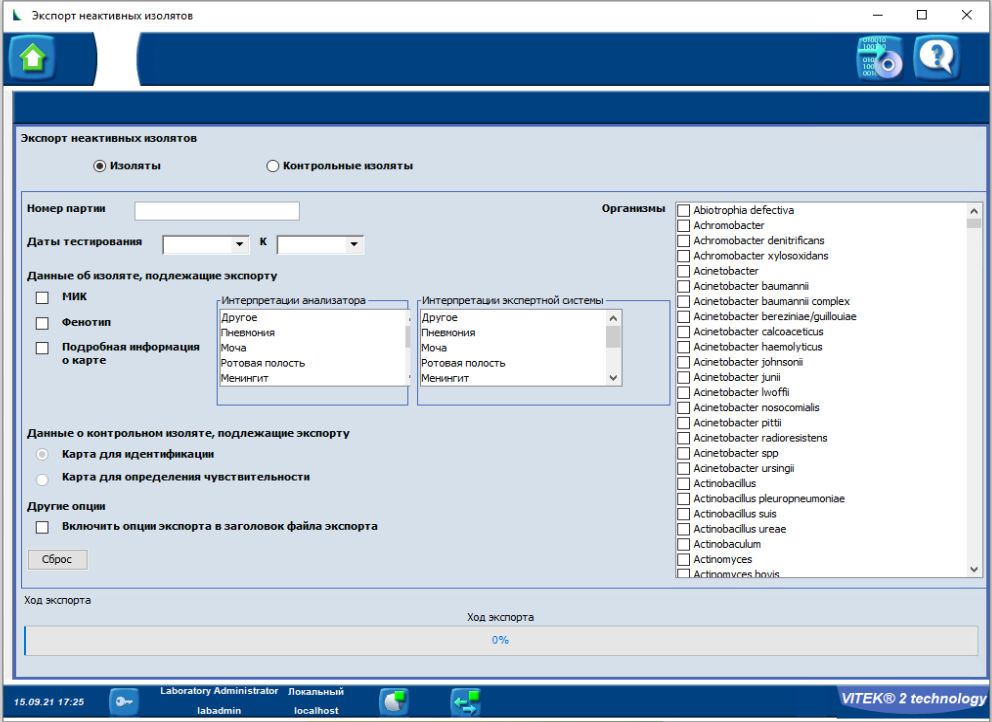
In the new window, click Yes.
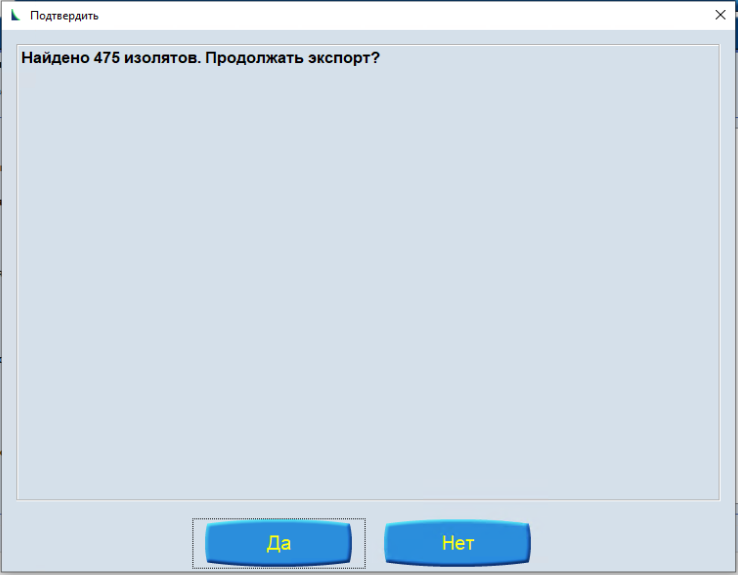
In the dialog box, select a location and enter a file name. Press Export inactive isolates.
Wait for the “Export over” message.
The resulting file must be passed through the converter to AMRcloud.
Online Converter
Preparation
The file with isolates from VITEK 2 system is a text file (* .txt), with a comma (,) field separator.
If you need to convert the file, follow the link https://public.amrcloud.net/vitek.
Uploading a File
To upload the file, click on the Choose file button and select your VITEK 2 file.
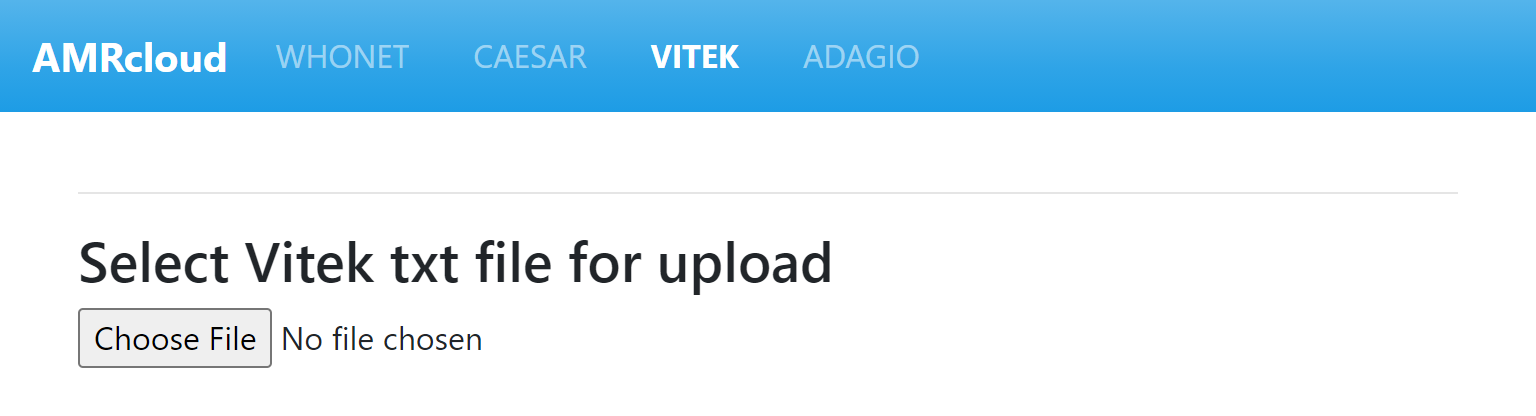
The file upload will start automatically. When the upload is complete, the system will check the contents of the file, try to read it, and display a preview on the screen. If the upload is successful, you will see the following.

If you see the table and the Convert button, then the file was successfully recognized by the system.
Converting a File
The file converting process consists in replacing the VITEK 2 antibiotic and organisms codes with AMRcloud codes.
As a result, the following columns are added to the table:
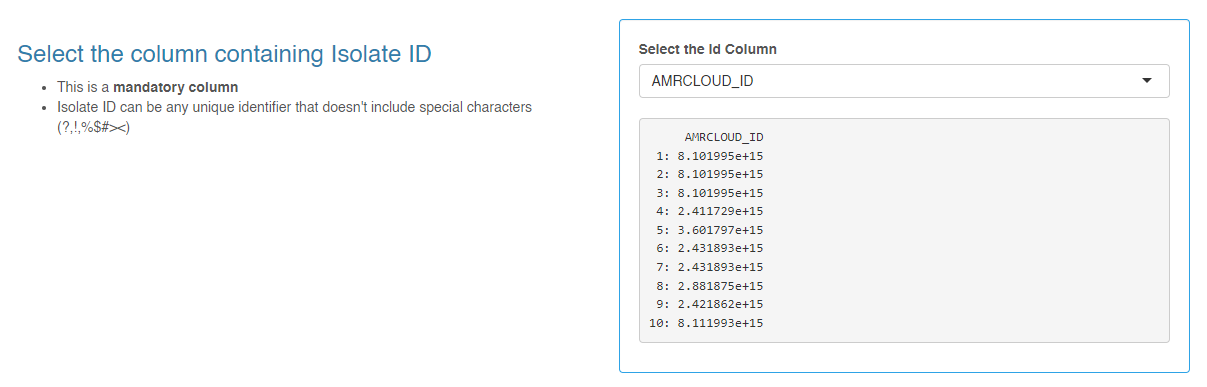
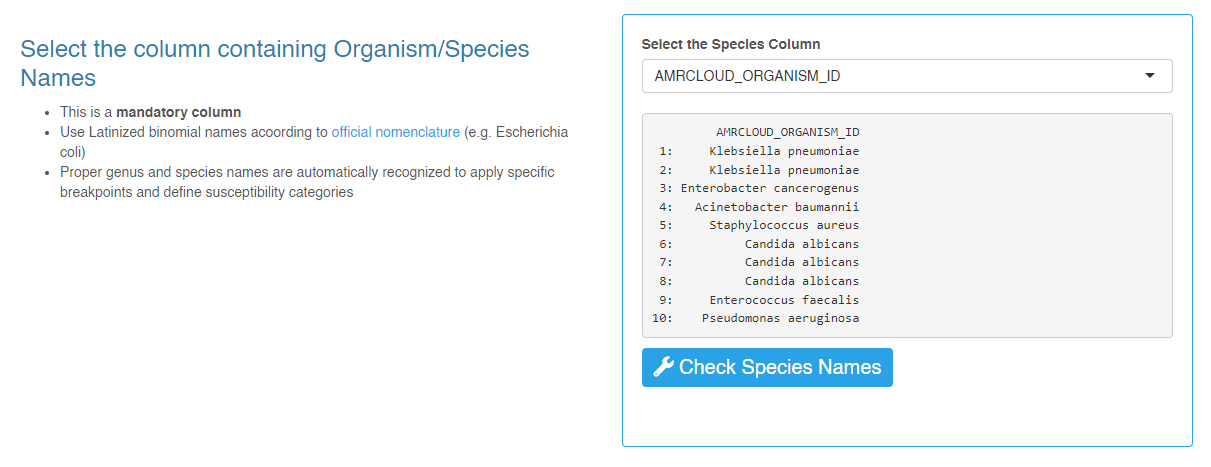
AMRCLOUD_ID- auto-generated unique row identifierAMRCLOUD_ORGANISM_ID- recoded names of microorganism species that allow AMRcloud to interpretations sensitivity data correct
- For item Select the column containing Isolate ID set
AMRCLOUD_ID - For item Select the column containing Organism/Species Names set
AMRCLOUD_ORGANISM_ID
Additionally, the values of several columns of resistance markers are combined into one column, starting with the code AMRCLOUD_MARKER_. The converter also removes columns with personal data - PATIENT NAME, PATIENT ID, PATIENT LOCATION, LAB ID.
After successfully uploading the file, click the Convert button. At the end of the converting process, a report on the performed manipulations will appear on the page. The report contains a list of codes for antibiotics and organisms that have been changed, as well as warning messages to look out for.
If errors occurred while analyzing the file, further converting is impossible. You need to fix the file.
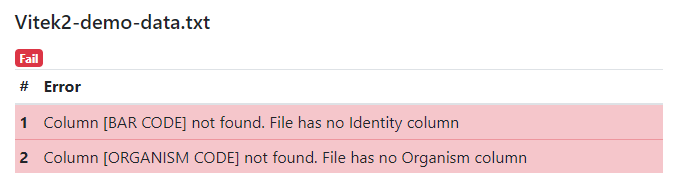
Possible errors:
- Error Column [BAR CODE] not found. File has no Identity column means that the BAR CODE column with unique isolate ID could not be found. You need to add it to the source file, or rename the existing column with the identifier.
- Error Column [ORGANISM CODE] not found. File has no Organism column means that the ORGANISM CODE column with the isolate microorganism ID could not be found. You need to add it to the source file, or rename the existing column.
There are the following types of warnings:
- Warning Column [AM-AMPICILLIN] looks like antibiotic, but has some non-MIC values means that the converter detects any non-numeric values in the antibiotic column. In this case, the column name is still converted to the AMRcloud format, the values remain unchanged.
- Warning Column AMRCLOUD_MARKER_AMINOGLYCOSIDE added means that the converter has detected columns with Aminoglycosides values and has created a new column with merged values for aminoglycosides.
- The warning Isolate with [BAR CODE] = 1234567890 has 2 rows means that the converter has detected that an isolate with an unique identifier BAR CODE occurs multiple times in the file. It is recommended to exclude duplicates from the analysis.
- The Column [AM-AMPICILLIN] has no values warning is displayed when no values are found in the antibiotic column.
- The warning Columns with identical names have been enumerated is displayed when the antibiotic column names are replaced with the same names. In this case, the columns are numbered in order.
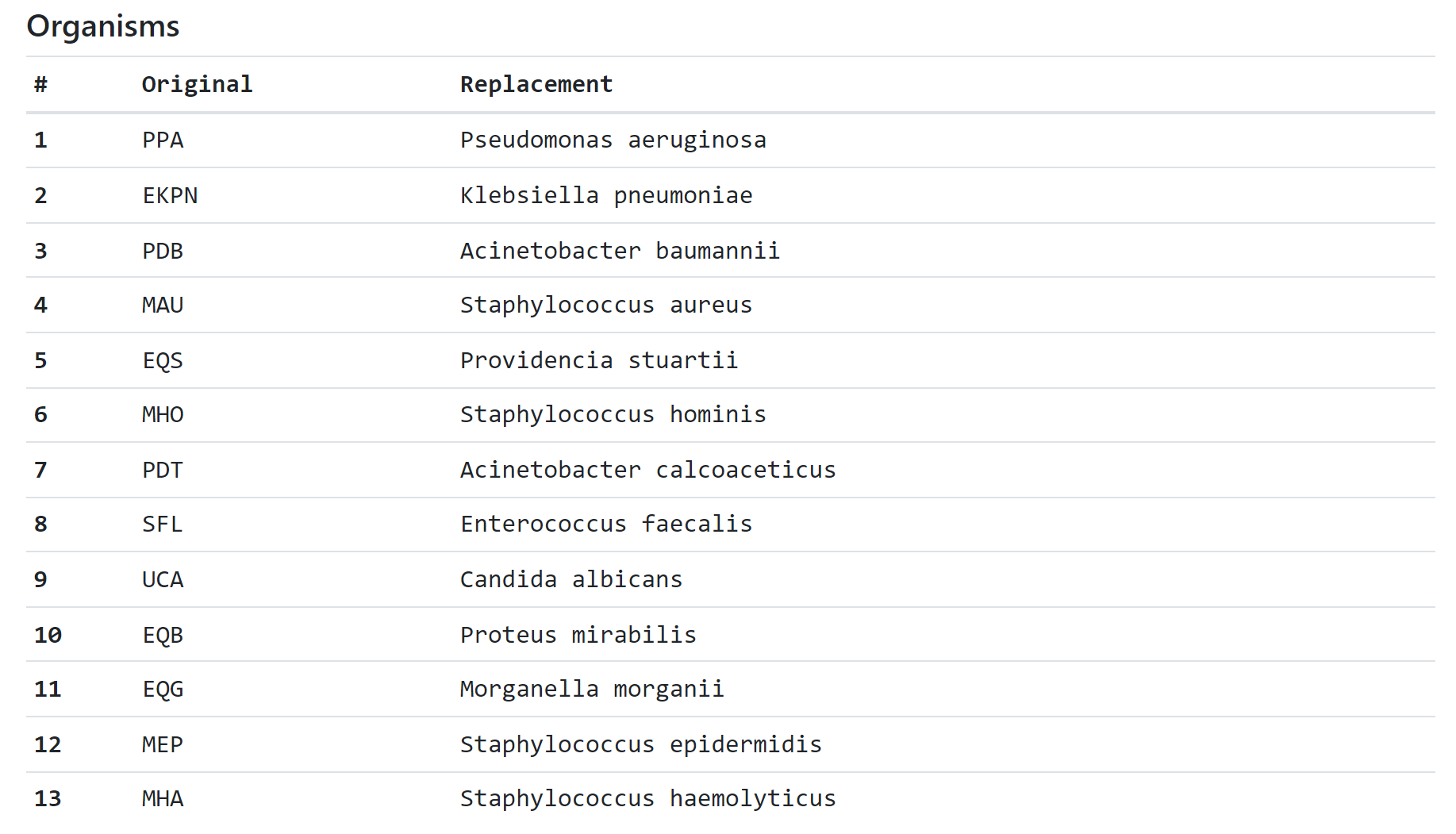
- The No translation has been found for organism [CODE] warning is displayed when the converter was unable to match the organism code from the ORGANISM CODE column with the dictionary. In this case, it is recommended to change the name of this microorganism manually.


Downloading a File
To get the converted file, click the Download button.
You can use this file to create projects and datasets in AMRcloud. The basic steps of uploading data to the AMRcloud online platform are described in the AMRcloud Data Import Tutorial.
Offline Converter
It is possible to convert data from VITEK 2 format to AMRcloud format without using the Internet. To do this, you need to use an offline converter.
Preparation
The file with isolates from VITEK 2 system is a text file (* .txt), with a comma (,) field separator.
Installation
To install the “AMRcloud Vitek Converter”, you should download it from the link.
If .NET Framework 4 is not installed on the computer, the installer will offer to download it. By default, the program is installed in the Documents\AMRsolution\AMRcloud Vitek Converter\ folder.
Working
The program window after launch looks like this.
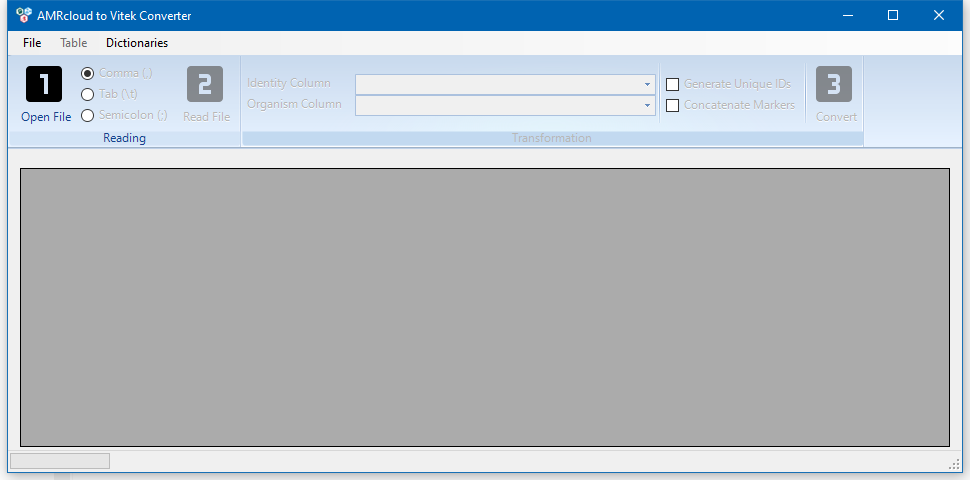
To select a VITEK file, press the Open File button, or select the File → Open Vitek File menu.
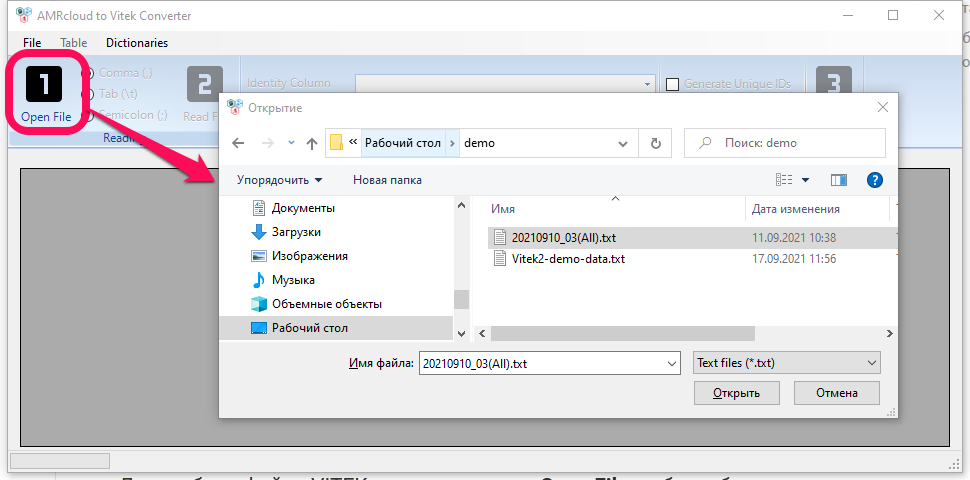
After selecting the file, press the Read File button or File → Read Vitek File menu. A table should appear. If the table does not appear, the file formatting may differ from the standard and you need to select a different type of separator. The separator type switch is located between the Open File and Read File buttons. The default is comma separator.
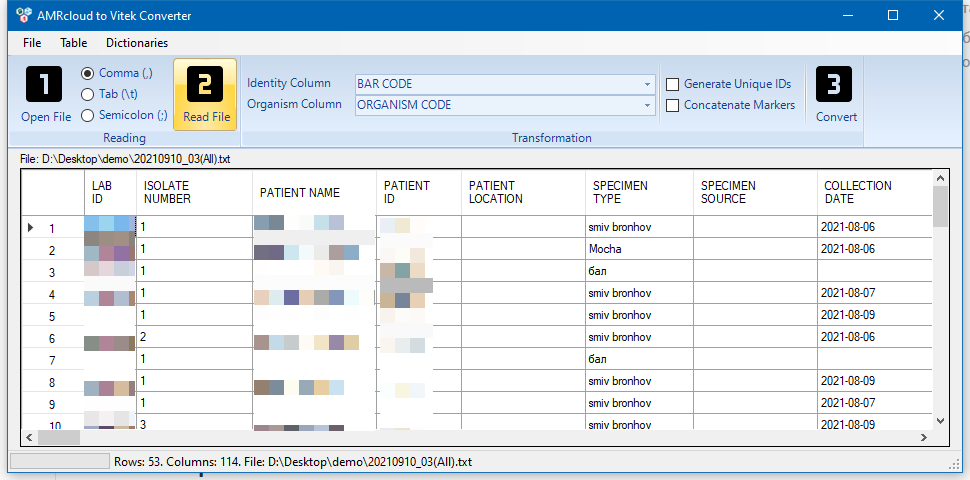
Next, you need to specify which columns contain the isolate ID and organism code. By default, it is assumed that the identifier is stored in the BAR CODE column, and the organism code in the ORGANISM CODE column. These values do not need to be changed.
If you wish, you can set additional transformation parameters:
- The Generate Unique IDs checkbox allows you to add a new column
AMRCLOUD_IDto the file with a new generated unique ID. - The Concatenate Markers checkbox allows you to concatenate the values of several columns of resistance markers into one column, starting with the code
AMRCLOUD_MARKER_.
As a result, the following columns are added to the table:
AMRCLOUD_ID- auto-generated unique row identifierAMRCLOUD_ORGANISM_ID- recoded names of microorganism species that allow AMRcloud to interpretations sensitivity data correctAMRCLOUD_MARKER_- values of resistance markers
- For item Select the column containing Isolate ID set
AMRCLOUD_ID - For item Select the column containing Organism/Species Names set
AMRCLOUD_ORGANISM_ID


To start the converting process, click the Convert button.
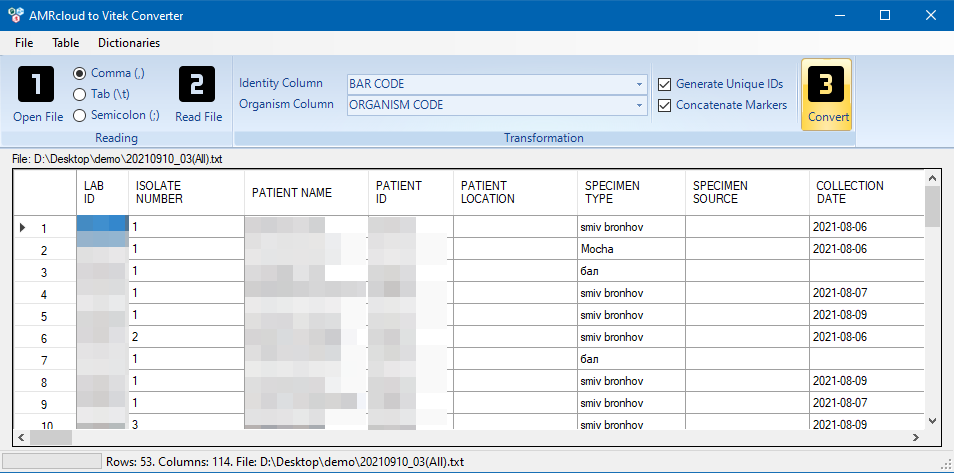
At the end of the converting process, a report on the performed manipulations will appear on the page. The report contains a list of codes for antibiotics and organisms that have been changed, as well as warning messages to look out for.
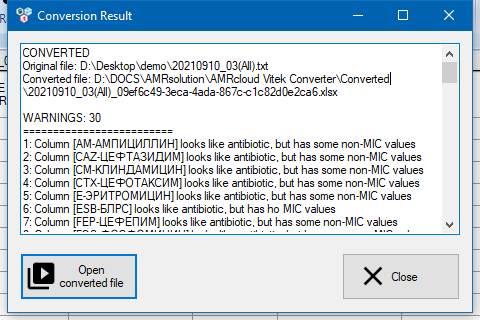
By default, the converted file is saved to the Documents\AMRsolution\AMRcloud Vitek Converter\Converted folder. You can open the converted file using the Open converted file button.
Possible errors:
- Error Column [BAR CODE] not found. File has no Identity column means that the BAR CODE column with unique isolate ID could not be found. You need to add it to the source file, or rename the existing column with the identifier.
- Error Column [ORGANISM CODE] not found. File has no Organism column means that the ORGANISM CODE column with the isolate microorganism ID could not be found. You need to add it to the source file, or rename the existing column.
There are the following types of warnings:
- Warning Column [AM-AMPICILLIN] looks like antibiotic, but has some non-MIC values means that the converter detects any non-numeric values in the antibiotic column. In this case, the column name is still converted to the AMRcloud format, the values remain unchanged.
- Warning Column AMRCLOUD_MARKER_AMINOGLYCOSIDE added means that the converter has detected columns with Aminoglycosides values and has created a new column with merged values for aminoglycosides.
- The warning Isolate with [BAR CODE] = 1234567890 has 2 rows means that the converter has detected that an isolate with an unique identifier BAR CODE occurs multiple times in the file. It is recommended to exclude duplicates from the analysis.
- The Column [AM-AMPICILLIN] has no values warning is displayed when no values are found in the antibiotic column.
- The warning Columns with identical names have been enumerated is displayed when the antibiotic column names are replaced with the same names. In this case, the columns are numbered in order.
- The No translation has been found for organism [CODE] warning is displayed when the converter was unable to match the organism code from the ORGANISM CODE column with the dictionary. In this case, it is recommended to change the name of this microorganism manually.
Dictionaries
The program contains pre-installed reference dictionaries of antibiotics and organisms, according to which the converting is carried out. If necessary, you can make changes to them using the Dicitonaries menu.
Dictionaries are subdivided into global and user’s. Global dictionaries contain predefined standard converting values. Custom references can override global codes. Therefore, it is recommended to make changes specifically to custom dictionaries.
Let’s select the Dictionaries → User’s organisms menu.
The dictionary window looks like this.
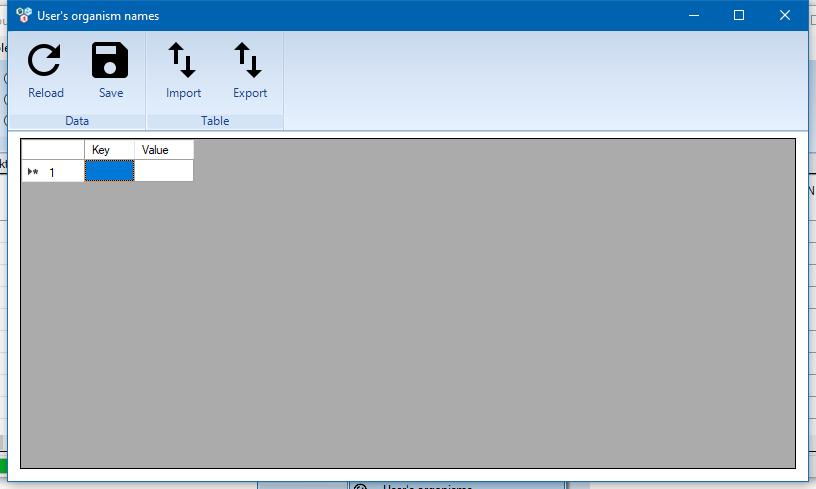
To enter the data, just double-click on a cell in the table and enter the text. Confirmation of the input is carried out with the Enter key. New entries are added using the line marked with an asterisk. You can save the changes using the Save button. The Reload button allows you to reload the table again, thereby resetting unsaved changes.
The Import button allows you to simplify the dictionary filling process. When you click the button, a text box appears where you can paste a table with codes from Excel. By pressing the OK button, they will be added to the dictionary.
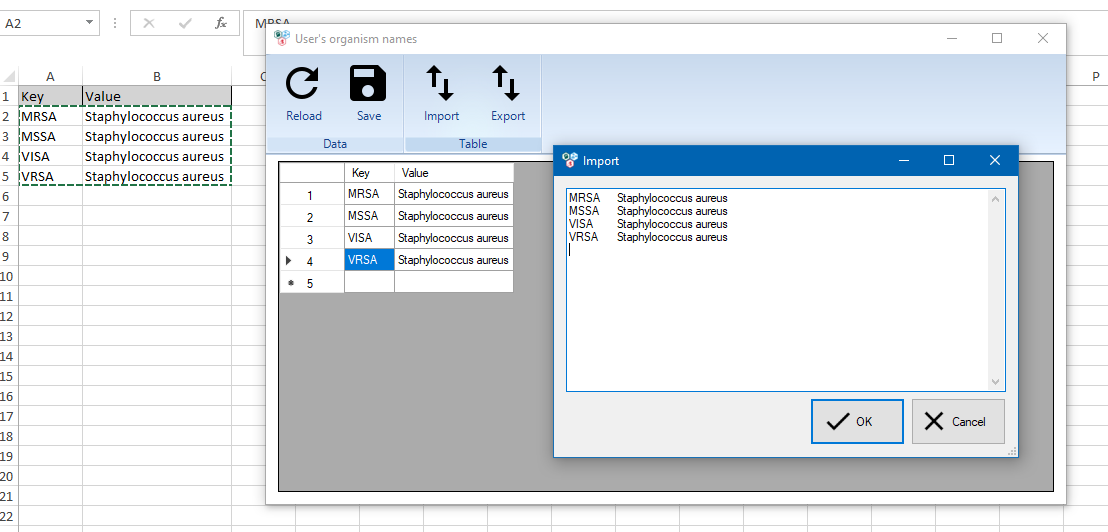
The Export button allows you to save the dictionary values to an Excel file.
Complex antibiotics replacement and Custom antibiotics replacement dictionaries
The peculiarity of these dictionaries is that they contain rules for converting MIC values for combined antibiotics.
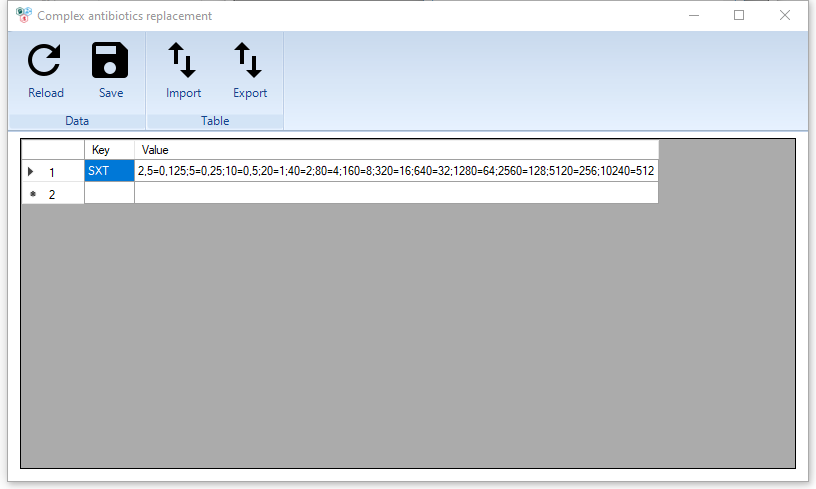
To describe such a converting, it is necessary to write pairs of values through the symbol (=), the comma is used as a separator of the pairs.
The figure shows an example for an SXT-Trimethoprim/sulfamethoxazole antibiotic. The following reference data were used for the dictionary.
| Vitek | trimethoprim, mg/L | sulfamethoxasole, mg/L |
|---|---|---|
| 2,5 | 0,125 | 2,375 |
| 5 | 0,25 | 4,75 |
| 10 | 0,5 | 9,5 |
| 20 | 1 | 19 |
| 40 | 2 | 38 |
| 80 | 4 | 76 |
| 160 | 8 | 152 |
| 320 | 16 | 304 |
| 640 | 32 | 608 |
| 1280 | 64 | 1216 |
| 2560 | 128 | 2432 |
| 5120 | 256 | 4864 |
| 10240 | 512 | 9728 |